Innovation & Workflow Optimization
I naturally take on optimizer/fixer roles within my groups, applying creativity and precision to enhance research tools and outcomes. To bypass complex issues I had multiple times found an innovative solution for my colleagues and for myself.
- Kymograph Analysis Pipeline (Open source)

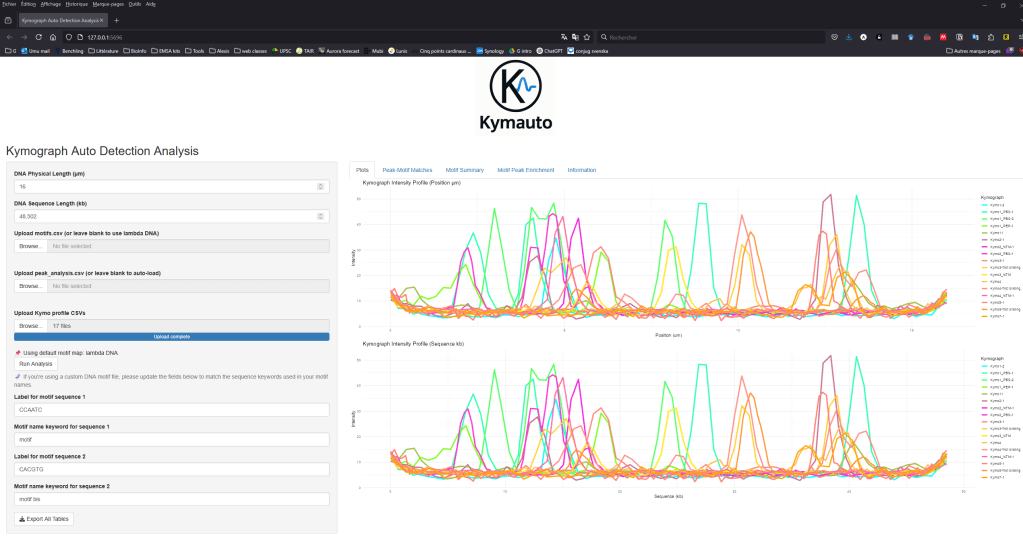
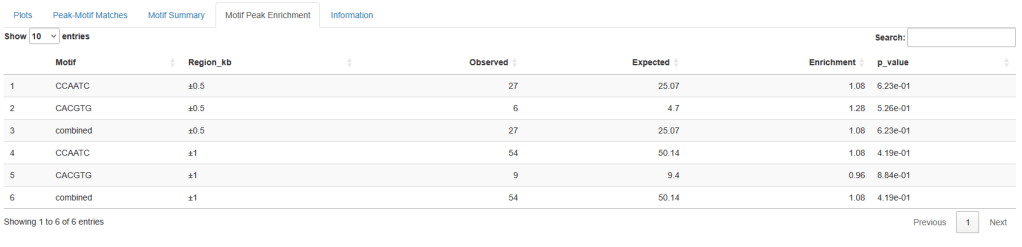
In my current group, we perform single-molecule interaction using the C-Trap optical tweezers technology from Lumicks. We are interested in the protein/DNA interaction. The company can supply us with special DNA handles that are labelled and are used to position precisely DNA regions along the optical tweezers. They act like beacons for us to know where interaction happens. Lumicks also provides an analysis basic software. Unfortunately, those handles are costly, very costly and not provided for many reactions and the basic analysis from the software isn’t enough for our needs. To bypass these two limiting costs we tried to work with lambda DNA, cheaper, without labelling but the positioning of interactions were difficult and not precise. I decided to create a tool using the kymographs from the C-Trap to position peaks intensity over a map of the lambda DNA (or other custom DNA maps) based on specific motifs (or custom motifs). This freed us from using Lumicks’ handles and we can analyse the interaction-peak positioning without paying for an upgraded license. Kymauto is a pipeline using ImageJ and R shiny apps (need to be pre-installed on your device), it is user-friendly : users don’t need any programming knowledge. It runs the interface with your own favorite web browser. It can analyse peak positioning, analyse your motifs specificity and statistical motif/peaks enrichment to extract any biological meaning from our experiment. Kymauto makes us save money, time, and release us from the help of a busy bioinformatician.
- “No-code” solutions for collaborators
I have at heart to find limiting bottlenecks of my collaborators to unlock valuable time for their research instead of repetitive “excel” tasks or modifying old scripts to analyse new data. I put efforts in finding fast, easy-to-use tools and workflows for data treatment, no matter its source (tables or pictures). KNIME is an open software that permitted me to create workflows such as a RT-qPCR analyser from machine output to plots not using one line of code or clics on an excel sheet. This is a plus if you can’t invest resources in training newly hired how to code basic scripts while maintaining data analysis quality and trust.
- Synthetic Protein Engineering in Yeast

Multicopper oxidases (MCOs) can be tricky proteins when it comes to measuring their activity. As it would have became an entire project by itself and with the clock ticking during my PhD, I designed and expressed a chimeric protein combining Arabidopsis and yeast domains to measure Multicopper Oxidase activity using mitochondrial oxygen consumption assays. With this technology I could make the measurements more efficient, simple and reproducible (see Publications section).
- High-Precision Seedling Phenotyping
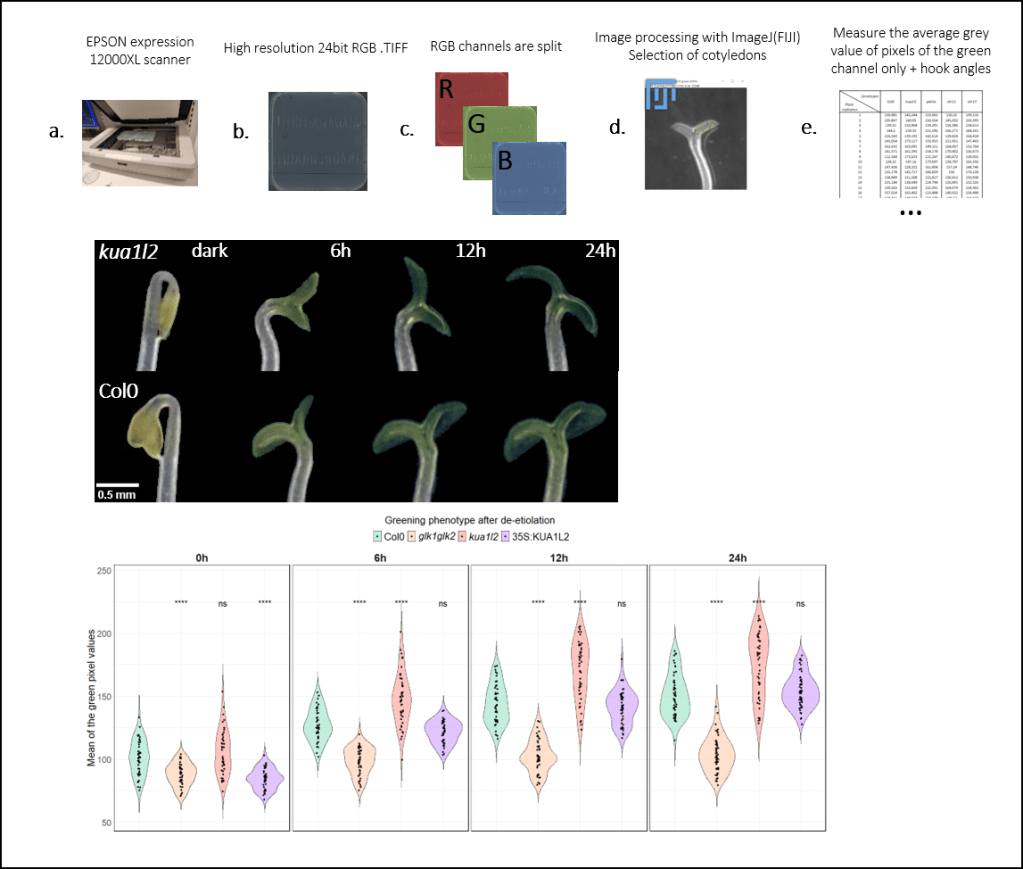
When I arrived in UPSC, my group were phenotyping early greening phenotype of plant using cameras in the greenhouse. It was nearly impossible to reproduce light conditions and resolution over time, leading to losing time and making color comparison untrustable. In my project, I had to screen dozens of mutants for ealy greening phenotype, I had to find a way to make it fast, high throughput, cheap, and reproducible to be useful for everyone. I designed a process using simple scanners coupled with ImageJ picture treatment software to perform this fast screen. With this fast method I could upscale the number of mutants and individuals and could show significant and repetable results. My group benefited from this new method and collaborators started to implement it in their own process (e.g. measure anthocyanin levels).
